Profiles¶
yt allows for data container objects to be binned up into profiles along some dimension defined by a field. These can be in 1D, 2D, or 3D. YT reproduces this functionality via the YTProfile type:
function YTProfile(data_source::DataContainer, bin_fields, fields;
n_bins=64, extrema=nothing, logs=nothing,
units=nothing, weight_field="cell_mass",
accumulation=false, fractional=false)
YTProfile takes the following arguments:
- data_source: The DataContainer object containing the data from which the profile is to be created.
- bin_fields: An Array of field names over which the binning will occur. The number of fields decides whether or not this will be a 1D, 2D, or 3D profile. If a single field string is given, it assumed to be 1D.
- fields: A single field or list of fields to be binned.
- n_bins: A single integer or tuple of 2 or 3 integers, to determine the number of bins along each dimension.
- extrema: A dictionary of tuples (with the field names as the keys) that determine the maximum and minimum of the bin ranges, e.g. [“density”=>(1.0e-30, 1.0e-25)]. If a field’s extrema are not specified, the extrema of the field in the data_source are assumed. The extrema are assumed to be in the units of the field in the units argument unless it is not specified, otherwise they are in the field’s default units.
- logs: A dictionary of tuples (with the field names as the keys) that determine whether the bins are in logspace or linear space, e.g. [“radius”=>false]. If not set, the take_log attribute for the field determines this.
- units: A dictionary of tuples (with the field names as the keys) that determine the units of the field. If not set then the default units for the field are used.
- weight_field: The field to weight the binned fields by when binning. Can be a field name or nothing, to produce an unweighted profile. "cell_mass" is the default.
- accumulation: If true, the profile values for a bin n are the cumulative sum of all the values from bin 1 to n. If the profile is 2D or 3D, an Array of values can be given to control the summation in each dimension independently.
- fractional: If true, the profile values are divided by the sum of all of the values.
For example, to construct a 1D radial profile from a Sphere, with the bins in linear space and with the units of the radius in kpc:
julia> sp = YT.Sphere(ds, "max", (1.0,"Mpc"))
julia> units=["radius"=>"kpc"]
julia> logs=["radius"=>false]
julia> fields=["density","temperature"]
julia> profile = YT.YTProfile(sp, "radius", fields, n_bins=100, units=units, logs=logs)
The bin_fields can be accessed from the YTProfile object as attributes, e.g.:
julia> profile.x
100-element YTArray (kpc):
4.99991
14.9997
24.9995
34.9993
44.9992
54.999
64.9988
74.9986
84.9984
94.9982
104.998
114.998
124.998
⋮
884.983
894.983
904.983
914.983
924.983
934.982
944.982
954.982
964.982
974.982
984.981
994.981
where the attributes x, y, and z correspond to the bin_fields of 1D, 2D, and 3D profiles. The fields of the profile are accessed in the same way as DataContainers:
julia> profile["temperature"]
100-element YTArray (K):
4.78287e7
4.78144e7
5.55494e7
5.98079e7
6.20128e7
6.41538e7
6.73181e7
7.28897e7
7.67484e7
7.6859e7
7.65575e7
7.60974e7
7.55863e7
⋮
5.15882e7
5.16148e7
5.15205e7
5.15374e7
5.15363e7
5.17031e7
5.15198e7
5.1652e7
5.16727e7
5.17993e7
5.18381e7
5.1944e7
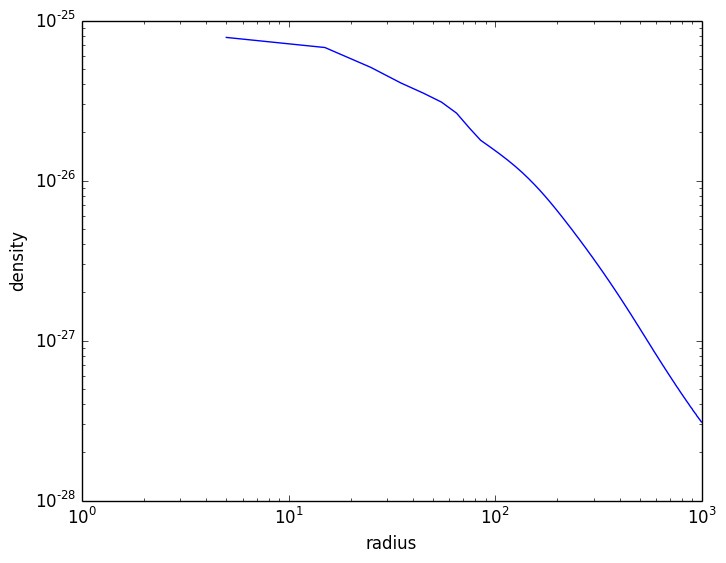
The resulting profile data can be plotted with a plotting program like PyPlot or Winston:
julia> using PyPlot
julia> plot(profile.x.value, profile["density"].value)
julia> xscale("log")
julia> yscale("log")
julia> xlabel("radius")
julia> ylabel("density")
The variance of a given field can be computed as well:
julia> YT.variance(profile, "density")
100-element YTArray (g/cm**3):
8.88606e-27
1.00439e-26
1.05204e-26
7.17655e-27
7.2972e-27
8.29273e-27
7.97938e-27
5.74176e-27
3.73228e-27
3.28493e-27
2.91421e-27
2.58537e-27
2.27903e-27
⋮
1.28528e-28
1.28161e-28
1.25986e-28
1.25587e-28
1.24444e-28
1.24095e-28
1.21943e-28
1.21903e-28
1.20435e-28
1.20124e-28
1.18991e-28
1.18264e-28
The units of the bin_fields can be changed using one of the set_x_unit, set_y_unit, or set_z_unit methods:
julia> YT.set_x_unit(profile, "Mpc")
julia> profile.x
100-element YTArray (Mpc):
0.00499991
0.0149997
0.0249995
0.0349993
0.0449992
0.054999
0.0649988
0.0749986
0.0849984
0.0949982
0.104998
0.114998
0.124998
⋮
0.884983
0.894983
0.904983
0.914983
0.924983
0.934982
0.944982
0.954982
0.964982
0.974982
0.984981
0.994981
Similarly, the units of the fields can be changed with set_field_unit:
julia> YT.set_field_unit(profile, "density", "Msun/kpc**3")
julia> profile["density"]
100-element YTArray (Msun/kpc**3):
1.16056e6
1.00365e6
754309.0
602161.0
519959.0
457351.0
390482.0
315199.0
264284.0
238734.0
217263.0
198420.0
181572.0
⋮
5663.04
5563.15
5426.81
5324.81
5220.5
5140.14
5011.73
4930.84
4834.15
4757.82
4663.7
4584.25